The Sequence
We now have Pfizer's Omicron BA.1 ("Riltozinameran") genetic sequence, alongside other important information, thanks to a FOI request to the UK's regulatory body, the MHRA.
The FOI Request
On the 3rd of September 2022 Matt Cooper has submitted the following FOI request to the UKs Medicines and Healthcare Products Regulatory Agency:
Dear Medicines and Healthcare Products Regulatory Agency,
Please release the following documents relating to the product "Comirnaty Original/Omicron BA.1 (15/15 micrograms)/dose dispersion for injection COVID-19 mRNA Vaccine (nucleoside modified)".
1) The full nucleotide base sequence for "Riltozinameran".
2) The conditions attached to this Conditional Marketing Authorisation.
3) The Public Assessment Report for this approval.
4) The date and time the application was made.
5) The date and time the application was approved.
Please provide the documents for 1), 2) and 3) in PDF format if possible.
Yours faithfully,
Matt Cooper
The response
On the 4th of October 2022, the MHRA Customer Services, Medicines and Healthcare Products Regulatory Agency has replied to him, with two PDFs. I hereby combined the response so it would be easier to see:
PART ONE - STRUCTURE / Sequence
MHRA response:
1) The full nucleotide base sequence for "Riltozinameran".
Please find attached a copy of the nucleotide sequence and background information related to this sequence [file name: structure]. It will be noted that this sequence is issued unredacted, and that this represents a different approach to partially redacted document that was recently issued in response to (22/949), i.e. another of your requests that pertained to the Moderna bivalent COVID-19 vaccine. The difference in handling is detailed fully in our 22/949 response, but to briefly summarise—the divergent approach is partly justified by differences in the tools and proprietary status of software which the companies used to optimise the nucleotide sequence.
3.2.S.1.2. STRUCTURE, OMICRON (B.1.1.529) VARIANT
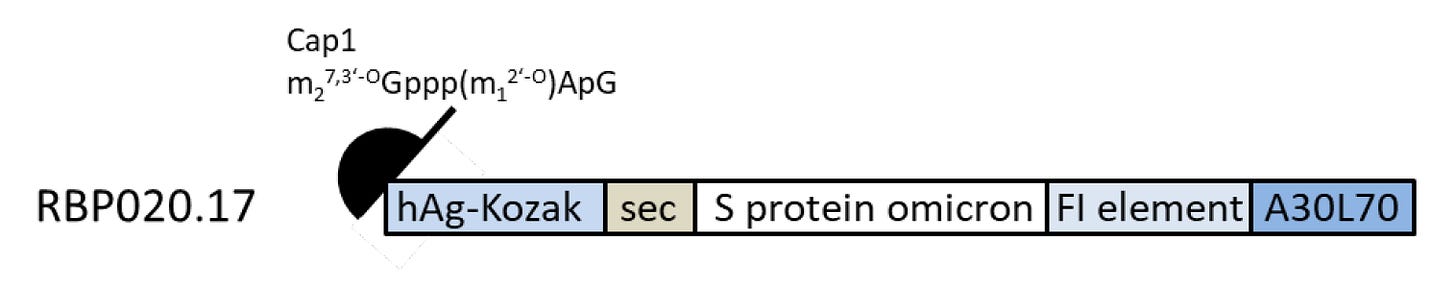
The active principle in each Omicron (B.1.1.529) variant drug substance (DS) is a single-stranded, 5'-capped mRNA that is translated into the respective protein (the encoded antigen). Figure 3.2.S.1.2-1 illustrates the general structure of the antigen-encoding RNA, which is determined by the respective nucleotide sequence of the DNA used as template for in vitro RNA transcription. In addition to the codon-optimized sequence encoding the antigen, the RNA contains common structural elements optimized for mediating high RNA stability and translational efficiency (5'-cap, 5'-UTR, 3'-UTR, poly(A) - tail; see below).
Furthermore, an intrinsic signal peptide (sec) is part of the antigen-encoding regions and is translated as N-terminal tag.
Figure 3.2.S.1.2-1. General structure of the Omicron (B.1.1.529) variant RNA
Schematic illustration of the general structure of the Omicron (B.1.1.529) drug substance with 5'-cap, 5'- and 3'-untranslated regions (hAg-Kozak and FI element, respectively), coding sequence for variant of concern and intrinsic signal peptide (sec) as well as poly(A)-tail (A30L70). Individual elements are not drawn to scale compared to their respective sequence lengths.
mRNA cap
A cap1 structure m2^(7,3’-O)Gppp(m1^(2’-O))ApG is utilized as specific capping structure at the 5′-end of the RNA drug substance (Figure 3.2.S.1.2-2).
Figure 3.2.S.1.2-2.
The cap1 structure (i.e., containing a 2′-O-methyl group on the penultimate nucleoside of the 5′-end of the RNA chain) is incorporated into the RNA drug substance by using a respective cap analog during in vitro transcription. For RNAs with modified uridine nucleotides, the cap1 structure is superior to other cap structures, since cap1 is not recognized by cellular factors such as IFIT1 and, thus, cap1-dependent translation is not inhibited by competition with eukaryotic translation initiation factor 4E. In the context of IFIT1 expression, mRNAs
with a cap1 structure give higher protein expression.
In addition, use of the cap1 structure leads to low amounts of uncapped transcripts. In general, the T7 Polymerase prefers a guanosine as priming nucleoside with the highest transcription efficiencies as compared to other starting nucleosides. Capping structures with a guanosine moiety compete with GTP for incorporation in the mRNA resulting in uncapped transcripts. The m2^(7,3’-O)Gppp(m1^(2’-O))ApG cap analog rescues transcription efficiency from templates starting with adenosines, because the ApG moiety of cap1 allows transcription initiation at the second position, a guanosine, thereby giving mainly capped mRNAs.
Modified Uridine
The RNA does not contain any uridines; instead of uridine the modified
N1-methylpseudouridine is used in RNA synthesis. Several reports have demonstrated that such a substitution often strongly enhances translation of in vitro transcribed mRNA sequences by reducing its immunogenicity5,6,7. Accordingly, the drug substance is synthesized in the presence of N1-methylpseudouridine triphosphate (m1ΨTP) instead of uridine triphosphate (UTP).
RNA sequence
The general sequence elements of the Omicron (B.1.1.529) variant drug substance, as depicted in Figure 3.2.S.1.2-1, are given below. The full sequence is given in Figure 3.2.S.1.2-3.
hAg-Kozak (nucleotides 2 to 54):
5'-UTR sequence of the human alpha-globin mRNA with an optimized ʻKozak sequenceʼ to increase translational efficiency.
Sec (nucleotides 55 to 102):
Sec corresponds to the intrinsic S1S2 protein signal peptide (sec), which guides translocation of the nascent polypeptide chain into the endoplasmic reticulum.
S protein omicron (nucleotides 103 to 3870):
Codon-optimized sequences encoding the respective antigen of SARS-CoV-2 protein has following point mutations/deletions (reference for numbering Genbank ID QHD43416.1): A67V, ΔHV69-70, T95I, G142D, ΔVYY143-145, ΔN211, L212I, R214_D215insEPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F, KV986-7PP.
FI element (nucleotides 3871 to 4165):
The 3′-UTR is a combination of two sequence elements derived from the “amino terminal enhancer of split” (AES) mRNA (called F) and the mitochondrial encoded 12S ribosomal RNA (called I). These were identified by an ex vivo selection process for sequences that confer RNA stability and augment total protein expression.
A30L70 (nucleotides 4166 to 4275):
A poly(A)-tail measuring 110 nucleotides in length, consisting of a stretch of 30 adenosine residues, followed by a 10 nucleotide linker sequence and another 70 adenosine residues designed to enhance RNA stability and translational efficiency in dendritic cells.
Figure 3.2.S.1.2-3. RNA nucleotide Sequence of the Omicron (B.1.1.529) drug substance
The nucleotide sequence 5’→3’is shown with individual sequence elements as indicated below.
GAGAAYAAAC YAGYAYYCYY CYGGYCCCCA CAGACYCAGA GAGAACCCGC 50
CACCAYGYYC GYGYYCCYGG YGCYGCYGCC YCYGGYGYCC AGCCAGYGYG 100
YGAACCYGAC CACCAGAACA CAGCYGCCYC CAGCCYACAC CAACAGCYYY 150
ACCAGAGGCG YGYACYACCC CGACAAGGYG YYCAGAYCCA GCGYGCYGCA 200
CYCYACCCAG GACCYGYYCC YGCCYYYCYY CAGCAACGYG ACCYGGYYCC 250
ACGYGAYCYC CGGCACCAAY GGCACCAAGA GAYYCGACAA CCCCGYGCYG 300
CCCYYCAACG ACGGGGYGYA CYYYGCCAGC AYCGAGAAGY CCAACAYCAY 350
CAGAGGCYGG AYCYYCGGCA CCACACYGGA CAGCAAGACC CAGAGCCYGC 400
YGAYCGYGAA CAACGCCACC AACGYGGYCA YCAAAGYGYG CGAGYYCCAG 450
YYCYGCAACG ACCCCYYCCY GGACCACAAG AACAACAAGA GCYGGAYGGA 500
AAGCGAGYYC CGGGYGYACA GCAGCGCCAA CAACYGCACC YYCGAGYACG 550
YGYCCCAGCC YYYCCYGAYG GACCYGGAAG GCAAGCAGGG CAACYYCAAG 600
AACCYGCGCG AGYYCGYGYY YAAGAACAYC GACGGCYACY YCAAGAYCYA 650
CAGCAAGCAC ACCCCYAYCA YCGYGAGAGA GCCCGAGGAY CYGCCYCAGG 700
GCYYCYCYGC YCYGGAACCC CYGGYGGAYC YGCCCAYCGG CAYCAACAYC 750
ACCCGGYYYC AGACACYGCY GGCCCYGCAC AGAAGCYACC YGACACCYGG 800
CGAYAGCAGC AGCGGAYGGA CAGCYGGYGC CGCCGCYYAC YAYGYGGGCY 850
ACCYGCAGCC YAGAACCYYC CYGCYGAAGY ACAACGAGAA CGGCACCAYC 900
ACCGACGCCG YGGAYYGYGC YCYGGAYCCY CYGAGCGAGA CAAAGYGCAC 950
CCYGAAGYCC YYCACCGYGG AAAAGGGCAY CYACCAGACC AGCAACYYCC 1000
GGGYGCAGCC CACCGAAYCC AYCGYGCGGY YCCCCAAYAY CACCAAYCYG 1050
YGCCCCYYCG ACGAGGYGYY CAAYGCCACC AGAYYCGCCY CYGYGYACGC 1100
CYGGAACCGG AAGCGGAYCA GCAAYYGCGY GGCCGACYAC YCCGYGCYGY 1150
ACAACCYGGC CCCCYYCYYC ACCYYCAAGY GCYACGGCGY GYCCCCYACC 1200
AAGCYGAACG ACCYGYGCYY CACAAACGYG YACGCCGACA GCYYCGYGAY 1250
CCGGGGAGAY GAAGYGCGGC AGAYYGCCCC YGGACAGACA GGCAACAYCG 1300
CCGACYACAA CYACAAGCYG CCCGACGACY YCACCGGCYG YGYGAYYGCC 1350
YGGAACAGCA ACAAGCYGGA CYCCAAAGYC AGCGGCAACY ACAAYYACCY 1400
GYACCGGCYG YYCCGGAAGY CCAAYCYGAA GCCCYYCGAG CGGGACAYCY 1450
CCACCGAGAY CYAYCAGGCC GGCAACAAGC CYYGYAACGG CGYGGCCGGC 1500
YYCAACYGCY ACYYCCCACY GCGGYCCYAC AGCYYYAGGC CCACAYACGG 1550
CGYGGGCCAC CAGCCCYACA GAGYGGYGGY GCYGAGCYYC GAACYGCYGC 1600
AYGCCCCYGC CACAGYGYGC GGCCCYAAGA AAAGCACCAA YCYCGYGAAG 1650
AACAAAYGCG YGAACYYCAA CYYCAACGGC CYGAAGGGCA CCGGCGYGCY 1700
GACAGAGAGC AACAAGAAGY YCCYGCCAYY CCAGCAGYYY GGCCGGGAYA 1750
YCGCCGAYAC CACAGACGCC GYYAGAGAYC CCCAGACACY GGAAAYCCYG 1800
GACAYCACCC CYYGCAGCYY CGGCGGAGYG YCYGYGAYCA CCCCYGGCAC 1850
CAACACCAGC AAYCAGGYGG CAGYGCYGYA CCAGGGCGYG AACYGYACCG 1900
AAGYGCCCGY GGCCAYYCAC GCCGAYCAGC YGACACCYAC AYGGCGGGYG 1950
YACYCCACCG GCAGCAAYGY GYYYCAGACC AGAGCCGGCY GYCYGAYCGG 2000
AGCCGAGYAC GYGAACAAYA GCYACGAGYG CGACAYCCCC AYCGGCGCYG 2050
GAAYCYGCGC CAGCYACCAG ACACAGACAA AGAGCCACCG GAGAGCCAGA 2100
AGCGYGGCCA GCCAGAGCAY CAYYGCCYAC ACAAYGYCYC YGGGCGCCGA 2150
GAACAGCGYG GCCYACYCCA ACAACYCYAY CGCYAYCCCC ACCAACYYCA 2200
CCAYCAGCGY GACCACAGAG AYCCYGCCYG YGYCCAYGAC CAAGACCAGC 2250
GYGGACYGCA CCAYGYACAY CYGCGGCGAY YCCACCGAGY GCYCCAACCY 2300
GCYGCYGCAG YACGGCAGCY YCYGCACCCA GCYGAAAAGA GCCCYGACAG 2350
GGAYCGCCGY GGAACAGGAC AAGAACACCC AAGAGGYGYY CGCCCAAGYG 2400
AAGCAGAYCY ACAAGACCCC YCCYAYCAAG YACYYCGGCG GCYYCAAYYY 2450
CAGCCAGAYY CYGCCCGAYC CYAGCAAGCC CAGCAAGCGG AGCYYCAYCG 2500
AGGACCYGCY GYYCAACAAA GYGACACYGG CCGACGCCGG CYYCAYCAAG 2550
CAGYAYGGCG AYYGYCYGGG CGACAYYGCC GCCAGGGAYC YGAYYYGCGC 2600
CCAGAAGYYY AAGGGACYGA CAGYGCYGCC YCCYCYGCYG ACCGAYGAGA 2650
YGAYCGCCCA GYACACAYCY GCCCYGCYGG CCGGCACAAY CACAAGCGGC 2700
YGGACAYYYG GAGCAGGCGC CGCYCYGCAG AYCCCCYYYG CYAYGCAGAY 2750
GGCCYACCGG YYCAACGGCA YCGGAGYGAC CCAGAAYGYG CYGYACGAGA 2800
ACCAGAAGCY GAYCGCCAAC CAGYYCAACA GCGCCAYCGG CAAGAYCCAG 2850
GACAGCCYGA GCAGCACAGC AAGCGCCCYG GGAAAGCYGC AGGACGYGGY 2900
CAACCACAAY GCCCAGGCAC YGAACACCCY GGYCAAGCAG CYGYCCYCCA 2950
AGYYCGGCGC CAYCAGCYCY GYGCYGAACG AYAYCYYCAG CAGACYGGAC 3000
CCYCCYGAGG CCGAGGYGCA GAYCGACAGA CYGAYCACAG GCAGACYGCA 3050
GAGCCYCCAG ACAYACGYGA CCCAGCAGCY GAYCAGAGCC GCCGAGAYYA 3100
GAGCCYCYGC CAAYCYGGCC GCCACCAAGA YGYCYGAGYG YGYGCYGGGC 3150
CAGAGCAAGA GAGYGGACYY YYGCGGCAAG GGCYACCACC YGAYGAGCYY 3200
CCCYCAGYCY GCCCCYCACG GCGYGGYGYY YCYGCACGYG ACAYAYGYGC 3250
CCGCYCAAGA GAAGAAYYYC ACCACCGCYC CAGCCAYCYG CCACGACGGC 3300
AAAGCCCACY YYCCYAGAGA AGGCGYGYYC GYGYCCAACG GCACCCAYYG 3350
GYYCGYGACA CAGCGGAACY YCYACGAGCC CCAGAYCAYC ACCACCGACA 3400
ACACCYYCGY GYCYGGCAAC YGCGACGYCG YGAYCGGCAY YGYGAACAAY 3450
ACCGYGYACG ACCCYCYGCA GCCCGAGCYG GACAGCYYCA AAGAGGAACY 3500
GGACAAGYAC YYYAAGAACC ACACAAGCCC CGACGYGGAC CYGGGCGAYA 3550
YCAGCGGAAY CAAYGCCAGC GYCGYGAACA YCCAGAAAGA GAYCGACCGG 3600
CYGAACGAGG YGGCCAAGAA YCYGAACGAG AGCCYGAYCG ACCYGCAAGA 3650
ACYGGGGAAG YACGAGCAGY ACAYCAAGYG GCCCYGGYAC AYCYGGCYGG 3700
GCYYYAYCGC CGGACYGAYY GCCAYCGYGA YGGYCACAAY CAYGCYGYGY 3750
YGCAYGACCA GCYGCYGYAG CYGCCYGAAG GGCYGYYGYA GCYGYGGCAG 3800
CYGCYGCAAG YYCGACGAGG ACGAYYCYGA GCCCGYGCYG AAGGGCGYGA 3850
AACYGCACYA CACAYGAYGA CYCGAGCYGG YACYGCAYGC ACGCAAYGCY 3900
AGCYGCCCCY YYCCCGYCCY GGGYACCCCG AGYCYCCCCC GACCYCGGGY 3950
CCCAGGYAYG CYCCCACCYC CACCYGCCCC ACYCACCACC YCYGCYAGYY 4000
CCAGACACCY CCCAAGCACG CAGCAAYGCA GCYCAAAACG CYYAGCCYAG 4050
CCACACCCCC ACGGGAAACA GCAGYGAYYA ACCYYYAGCA AYAAACGAAA 4100
GYYYAACYAA GCYAYACYAA CCCCAGGGYY GGYCAAYYYC GYGCCAGCCA 4150
CACCCYGGAG CYAGCAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAGCAYA 4200
YGACYAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 4250
AAAAAAAAAA AAAAAAAAAA AAAAA 4275
Sequence length: 4275, which includes G to denote the presence of the 5’-cap analog G: 1062 C: 1305 A: 1108 Y: 800 A = Adenine; C = Cytosine; G = Guanine; Y = N1-methylpseudouridine
The conditions attached to this Conditional Marketing Authorisation.
The conditions will be published appended to the public assessment report, however, we recognise that there is a public interest in the conditions & postauthorisation commitments and so we have provided these (see below). Please note, in our response to 22/949 we instead applied Section 22 (Information intended for future publication) to the conditions because the PAR corresponding to that request is expected to be published in the coming days.
This authorisation has the following post authorisation measure(s) which should be fulfilled by the date(s) shown if a date is specified:
1. The MAH should submit the 3 and 6 month post dose 4 immunogenicity and safety results in participants aged > 55 years from Study C4591031 Substudy.
E. Due date 31 January 2023
2. The MAH should submit the cellular immunogenicity results from study C4591031 Substudy.
E. Due date 31 March 2023
3. The MAH should submit the 1, 3 and 6 month post dose 4 immunogenicity and safety results in participants aged 18 to 55 years from Study C4591031 Substudy.
E. Due date 31 March 2023 (1 month data due by 31 October 2022)
4. The MAH should submit a standalone summary safety report for the Original / Omicron BA.I bivalent product, with the data lock point falling 3 months after the date of approval in Great Britain.
Due date 16 December 2022
5. Within one month of approval, the MAH must submit the following concerning Post-Authorisation Vaccine Effectiveness:
a. Confirmation that ongoing UK-based effectiveness study W1255886 will be promptly amended to include the collection of effectiveness data for the bivalent vaccine
b. An analysis of the feasibility and power of study W1255886 to generate robust results for the bivalent vaccine
c. Milestones for the provision of results for the bivalent vaccine
d. A full updated study protocol to reflect the investigation of the bivalent product
Due date 02 October 2022
6. Where there is a business need to manufacture Omicron (BA.I) circular plasmid DNA and linear DNA template at Pfizer Zagreb, the batch analysis data obtained from the first commercial batch from Pfizer Zagreb shall be provided.
7. Comparative accelerated stability data beüteen the prototype and bivalent vaccine drug product should be presented to ensure comparable stability profiles are seen for the vaccines.
The Public Assessment Report for this approval
The full PAR is exempt under Section 22 of the FOIA (Information intended for future publication), the PAR is due to be published in the coming few weeks. While there is a public interest in the PAR, we can see no advantage in providing a non-finalised version which is due to be published in a short-time scale.
Please note, the following information on the EMA website may also be of interest:
Comirnaty, INN-tozinameran, tozinameran/riltozinameran (europa.eu)
The date and time the application was made and approved
According to our records, the application for Comirnaty Original/Omicron BA.1 (15/15 micrograms)/dose Dispersion for Injection (PLGB 53632/0010) was received on 09 June 2022 and granted on 02 September 2022.
I’ll be sharing more on the topic soon… stay tuned…
Don’t forget to join my Telegram channel!
If you want to see the two PDFs provided to Matt as part of the FOI, you can find them here.
Love,
Ehden Biber


N501Y, Q493R,T478K and more in da house!
Enhanced binding to ACE2 also added for good measure in the transfection-soup. This will be getting interesting soon...
(e.g. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8811907/)
Thank you!
When ever I see a post from you, I think that this is the BRILLIANT PSEUDOURIDINE GUY.
But, alas, this material is so hard, way over my head.
I vouchsafe that even the Cliffs Notes would be too hard for me.